生命医科学研究科博士前期課程1年大崎和さん、情報計算科学生物(CBI)学会ベストポスター賞受賞!
2019.10.31
- TOPICS
- 研究
- 学生の活躍
- 教育
- 大学
生命情報科学研究室前期課程1年 大崎和さん(おおさき かず)、情報計算科学生物(CBI)学会ベストポスター賞受賞!
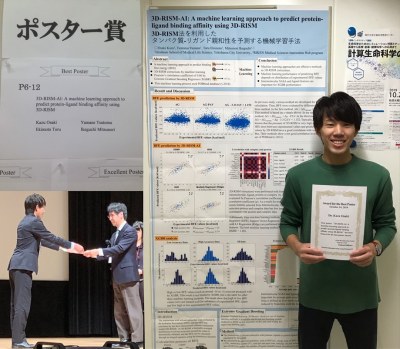
発表演題「3D-RISM-AI: A machine learning approach to predict protein-ligand binding affinity using 3D-RISM」
大学院生命医科学研究科博士前期課程1年の大崎和さん(指導教員:池口満徳教授)は、2019年10月22日(火)〜24日(木)にタワーホール船堀(東京都)で開催された情報計算科学生物(CBI)学会2019年大会においてポスター発表を行い、ベストポスター賞を受賞されました。
大崎和さんのコメント
この度は栄えある賞をいただき大変うれしく、光栄に思います。発表準備にあたり池口教授、及び研究室の皆様から熱心にご協力していただきました。心より感謝申し上げます。当学会中、様々な専門家の方々と議論を交わした時間は、私にとって非常に新鮮で刺激的でした。自分の研究について多角的な視点からアドバイスを頂き、自分の未熟さを思い知りながらも、研究により一層邁進していく気持ちが強くなりました。今回の受賞を励みに、今後も研究活動に注力していきたいと考えています。
指導教員の池口満徳教授のコメント
この4月に研究室に入ってきてから、10月までの短い期間でよくここまでの研究をまとめたと思います。様々なことを自分で調べながらどんどん研究を進める積極的な姿勢はとてもすばらしく、それが発表の質疑応答でもよく出ていたのではないかと思います。ぜひ、今後も、成果を論文にまとめるなど、研究を発展させていってください。
発表内容
Accurately predicting protein-ligand binding affinities is important in drug discovery, because the protein-ligand affinity is a primary criteria to evaluate stabilities for abundant protein-ligand complexes in virtual screening and/or docking simulation. The protein-ligand affinity is given by the (relative) binding free energy, and many computational methods have been developed using empirical scores and molecular dynamics simulations etc. Recently, the prediction of the affinity has been evaluated by machine learning in which intermolecular interactions between protein and ligand, structural features, and amino acid sequences were taken into account as descriptors [1].
In the binding free energy calculation, interactions of protein/ligand with solvent molecules are considered as an important factor to improve accuracy of the prediction. Therefore, we propose a machine learning approach taking into account the interactions with solvent molecules (3D-RISM-AI). The interactions with solvent molecules were evaluated by the hydration free energy (HFE), and the HFE was calculated by the three-dimensional reference interaction model (3D-RISM) [2, 3]. In 3D-RISM, a thermally averaged distribution of solvent molecules around the solute is theoretically obtained, and the HFE can be calculated through an integral equation using the distribution function. We calculated ~3800 of the HFE for protein-ligand complexes in the PDBbind database (v.2018) and used the data in the learning process. Whereas the binding free energies solely evaluated using 3D-RISM are not correlated with the experimental data, the binding free energies predicted using 3D-RISM-AI show a good correlation with the experimental values.
[1] Ain, Q. U. et al. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2015, 5, 405−424
[2] D. Beglov and B. Roux, J. Chem. Phys. 103, 360, 1995.
[3] D. Beglov and B. Roux, J. Chem. Phys. 104, 8678, 1996
In the binding free energy calculation, interactions of protein/ligand with solvent molecules are considered as an important factor to improve accuracy of the prediction. Therefore, we propose a machine learning approach taking into account the interactions with solvent molecules (3D-RISM-AI). The interactions with solvent molecules were evaluated by the hydration free energy (HFE), and the HFE was calculated by the three-dimensional reference interaction model (3D-RISM) [2, 3]. In 3D-RISM, a thermally averaged distribution of solvent molecules around the solute is theoretically obtained, and the HFE can be calculated through an integral equation using the distribution function. We calculated ~3800 of the HFE for protein-ligand complexes in the PDBbind database (v.2018) and used the data in the learning process. Whereas the binding free energies solely evaluated using 3D-RISM are not correlated with the experimental data, the binding free energies predicted using 3D-RISM-AI show a good correlation with the experimental values.
[1] Ain, Q. U. et al. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2015, 5, 405−424
[2] D. Beglov and B. Roux, J. Chem. Phys. 103, 360, 1995.
[3] D. Beglov and B. Roux, J. Chem. Phys. 104, 8678, 1996