横浜市立大学 生命医科学研究科
構造エピゲノム科学研究室
助教
小沼 剛 Ph.D.
Lab : B305
E-mail : konumax[at]yokohama-cu.ac.jp
[at]を@に変えて下さい
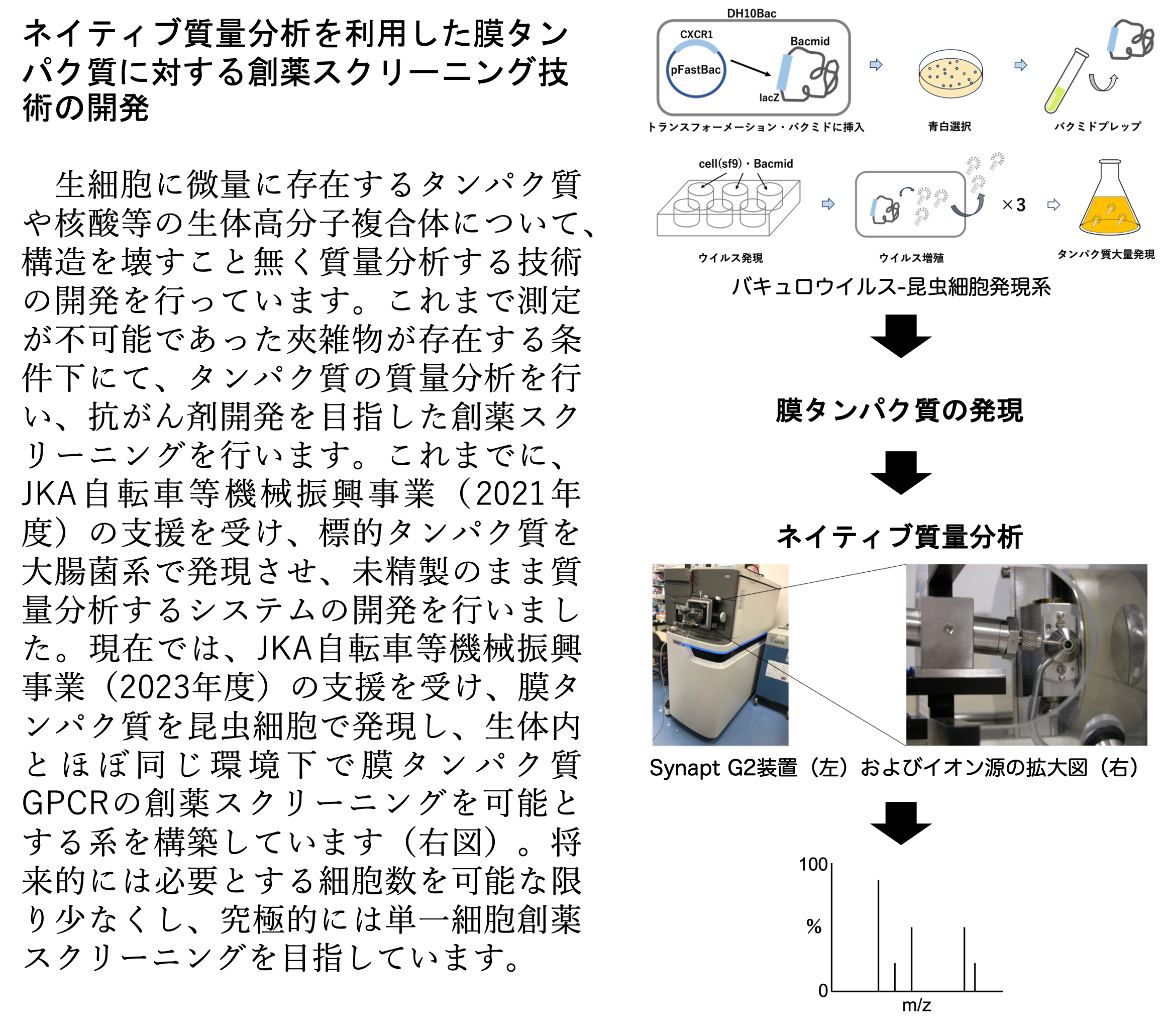
【研究内容】
核磁気共鳴(NMR)や質量分析(MS)、X線、クライオ電子顕微鏡を利用し、タンパク質や核酸などの生体分子の相互作用解析や立体構造解析を行い生命活動の基礎を研究する。また、がんなど重篤な疾病に関わるタンパク質について、その立体構造に基づいた抗がん剤の開発を行う。
【主要論文】
1. Konuma T, Takai T, Tsuchiya C, Nishida M, Hashiba M, Yamada Y, Nagadoi A, Chikaishi E, Akagi K, Akashi S, Yamazaki T, Akustu H, Ikegami T.
“NMR analysis of the homodimeric structure of a D-Ala-D-Ala metallopeptidase, VanX, from vancomycin-resistant bacteria”
Pro. Sci., 33, e5002 (2024)
2. #Suzuki N, #,*Konuma T, Ikegami T, *Akashi S
“Biophysical insights into the dimer formation of human sirtuin 2”
Pro. Sci., 33, e4994 (2024) (#Co-first author, *Co-corresponding author)
3. Konuma T, Zhou MM
“Distinct Histone H3 lysine 27 modifications dictate different outcomes of gene transcription”
J. Mol. Biol., 436, 168376 (2024)
4. Sayaka H, Imagawa E, Hirano D, Ikegami T, Oishi K, Konuma T
“1H, 13C and 15N backbone resonance assignments of hepatocyte nuclear factor-1-beta (HNF1β) POUS and POUHD”
Biomol. NMR Assign., 18, 59-63 (2024)
5. #Liu N, #Konuma T, Sharma R, Wang D, Zhao N, Cao L, Ju Y, Liu D, Wang S, Bosch A, Sun Y, Zhang S, Ji D, Nagatoishi S, Suzuki N, Kikuchi M, Wakamori M, Zhao CC, Ren C, Zhou J T, Xu Y, Meslamani J, Fu S, Umehara T, Tsumoto K, Akashi S, Zeng L, Roeder G R, Walsh J M, Zhang Q, Zhou MM
“Histone H3 lysine 27 crotonylation mediates gene transcriptional repression in chromatin”
Mol. Cell, 83, 2206-2221 (2023) Seleced as Featured Article (#Co-first author)
6. Konuma T, Kurita J, Ikegami T
“CPMG pulse sequence for relaxation dispersion that cancels artifacts independently of spin states”
J. Mag. Res., 352, 107489 (2023)
7. Azegami N, Taguchi R, Suzuki N, Sakata Y, *Konuma T, *Akashi S
“Native mass spectrometry of BRD4 bromodomains linked to a long disordered region”
Mass Spectrom. (Tokyo), 11, A0110 (2022) (*Co-corresponding author)
8. Hata K, Kobayashi N, Sugimura K, Qin W, Haxholli D, Chiba Y, Yoshimi S, Hayashi G, Onoda H, Ikegami T, Mulholland BC, Nishiyama A, Nakanishi M, Leonhardt H, *Konuma T, *Arita K
“Structural basis for the unique multifaceted interaction of DPPA3 with the UHRF1 PHD finger”
Nucleic. Acids. Res., 50, 12527-12542 (2022) (*Co-corresponding author)
9. Sakamoto W, Azegami N, Konuma T, Akashi S
“Single cell native mass spectrometry of human erythrocytes”
Anal. Chem., 93, 6583-6588 (2021)
10. Konuma T, Yu D, Zhao CC, Ju Y, Sharma R, Ren C, Zhang Q, Zhou MM, Zeng L
“Structural mechanism of the oxygenase JMD6 recognition by the extraterminal (ET) domain of BRD4”
Sci. Rep., 7, 16272-16281 (2017)
Researchmap
【研究助成】

【研究内容】